-
Notifications
You must be signed in to change notification settings - Fork 12
/
Copy pathREADME.Rmd
126 lines (81 loc) · 4.78 KB
/
README.Rmd
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
---
output: github_document
---
<!-- README.md is generated from README.Rmd. Please edit that file -->
# [CDMConnector](https://darwin-eu.github.io/CDMConnector/)
<!-- badges: start -->
[](https://lifecycle.r-lib.org/articles/stages.html#stable)
[](https://CRAN.R-project.org/package=CDMConnector)
[](https://app.codecov.io/gh/darwin-eu/CDMConnector?branch=main)
[](https://github.com/darwin-eu/CDMConnector/actions?query=workflow%3AR-CMD-check)
[](https://github.com/darwin-eu/CDMConnector/actions?query=workflow%3Aduckdb-test)
[](https://github.com/darwin-eu/CDMConnector/actions?query=workflow%3Apostgres-test)
[](https://github.com/darwin-eu/CDMConnector/actions?query=workflow%3Asqlserver-odbc-test)
[](https://github.com/darwin-eu/CDMConnector/actions?query=workflow%3Aredshift-test)
[](https://github.com/darwin-eu/CDMConnector/actions?query=workflow%3Asnowflake-odbc-test)
<!-- badges: end -->
> Are you using the [tidyverse](https://www.tidyverse.org/) with an OMOP Common Data Model?
>
> Interact with your CDM in a pipe-friendly way with CDMConnector.
>
> - Quickly connect to your CDM and start exploring.
> - Build data analysis pipelines using familiar dplyr verbs.
> - Easily extract subsets of CDM data from a database.
## Overview
CDMConnector introduces a single R object that represents an OMOP CDM relational database inspired by the [dm](https://dm.cynkra.com/), [DatabaseConnector](http://ohdsi.github.io/DatabaseConnector/), and [Andromeda](https://ohdsi.github.io/Andromeda/) packages. The cdm objects encapsulate references to [OMOP CDM tables](https://ohdsi.github.io/CommonDataModel/) in a remote RDBMS as well as metadata necessary for interacting with a CDM, allowing for dplyr style data analysis pipelines and interactive data exploration.
[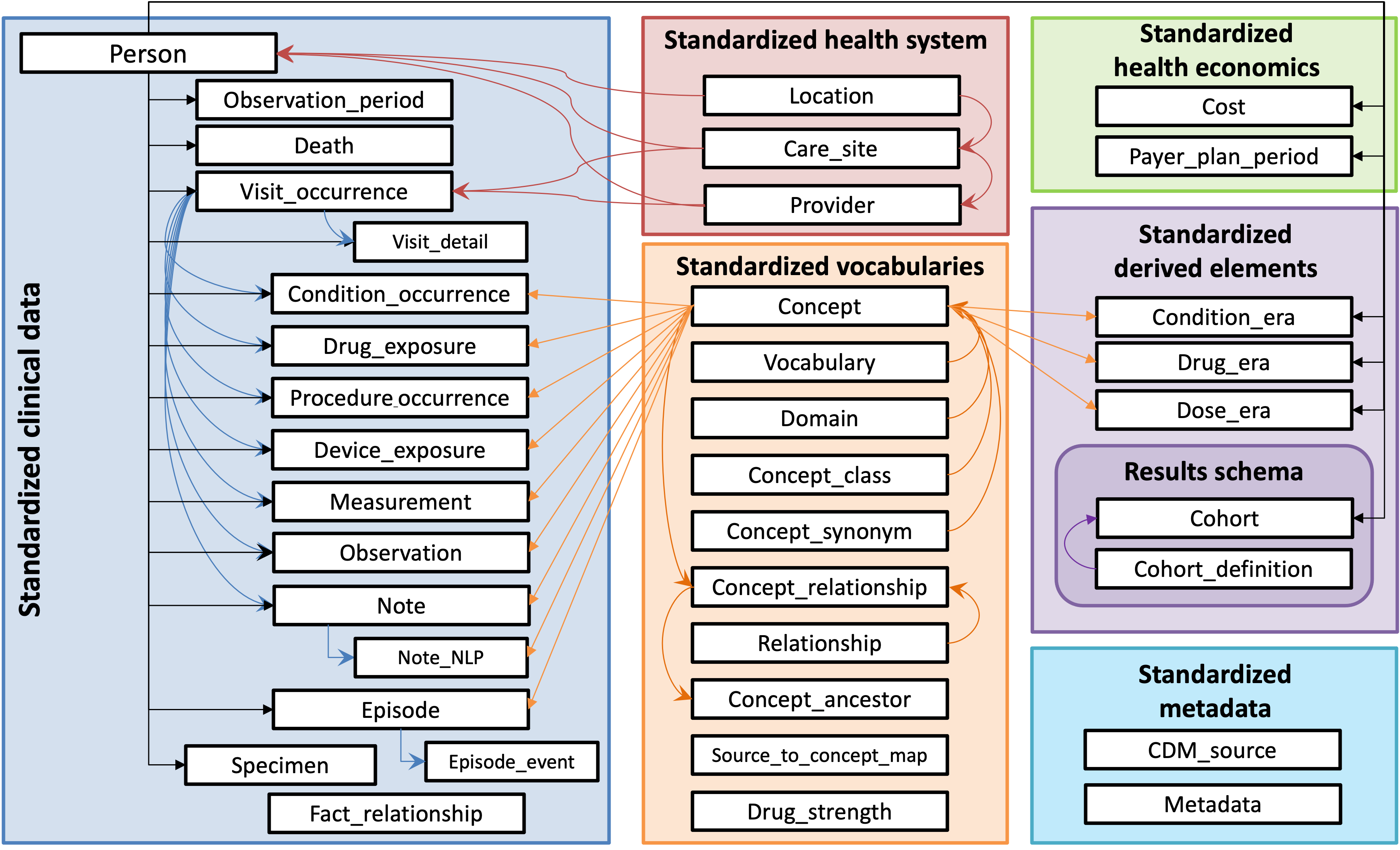](https://ohdsi.github.io/CommonDataModel/)
## Features
CDMConnector is meant to be the entry point for composable tidyverse style data analysis operations on an OMOP CDM.
A `cdm_reference` object behaves like a named list of tables.
- Quickly create a list of references to a subset of CDM tables
- Store connection information for later use inside functions
- Use any DBI driver back-end with the OMOP CDM
See Getting started for more details.
## Installation
CDMConnector can be installed from CRAN:
```r
install.packages("CDMConnector")
```
The development version can be installed from GitHub:
```r
# install.packages("devtools")
devtools::install_github("darwin-eu/CDMConnector")
```
## Usage
Create a cdm reference from any DBI connection to a database containing OMOP CDM tables. Use the cdm_schema argument to point to a particular schema in your database that contains your OMOP CDM tables and the write_schema to specify the schema where results tables can be created, and use cdm_name to provide a name for the database.
```{r}
library(CDMConnector)
con <- DBI::dbConnect(duckdb::duckdb(dbdir = eunomia_dir()))
cdm <- cdm_from_con(con = con,
cdm_schema = "main",
write_schema = "main",
cdm_name = "my_duckdb_database")
```
A `cdm_reference` is a named list of table references:
```{r, warning=FALSE, message=FALSE}
library(dplyr)
names(cdm)
```
Use dplyr verbs with the table references.
```{r}
cdm$person %>%
tally()
```
Compose operations with the pipe.
```{r}
cdm$condition_era %>%
left_join(cdm$concept, by = c("condition_concept_id" = "concept_id")) %>%
count(top_conditions = concept_name, sort = TRUE)
```
And much more besides. See vignettes for further explanations on how to create database connections, make a cdm reference, and start analysing your data.
```{r, include=FALSE}
cdmDisconnect(cdm)
```
## Getting help
If you encounter a clear bug, please file an issue with a minimal [reproducible example](https://reprex.tidyverse.org/) on [GitHub](https://github.com/darwin-eu/CDMConnector/issues).
## Citation
```{r, echo=FALSE, warning=FALSE}
citation("CDMConnector")
```
---
License: Apache 2.0